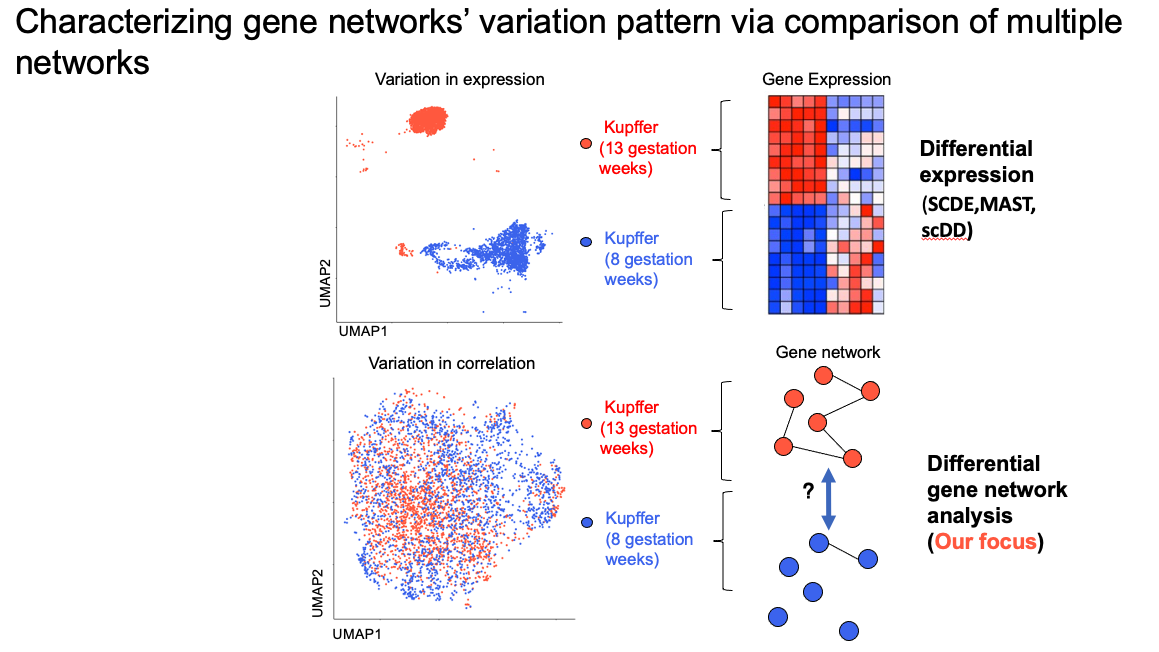
Introduction
Large fields of single transcriptomes study have extensively explored the effect of variation of genes’ mean expression. For example, In the left upper panel, 2 groups of Kupffer cells are presented. Their cell’s identity can be confirmed from genes which are differentially expressed in one cell group while lack of expression in another. However, the variation of genes’ expression is normally controlled by more complex regulatory mechanism such as gene pathways. In this work, what we are primarily interested in is to capture shifts and change of those gene coexpression networks (GCNs). (This project is in progress)
Autoregressive graph generation (2020)
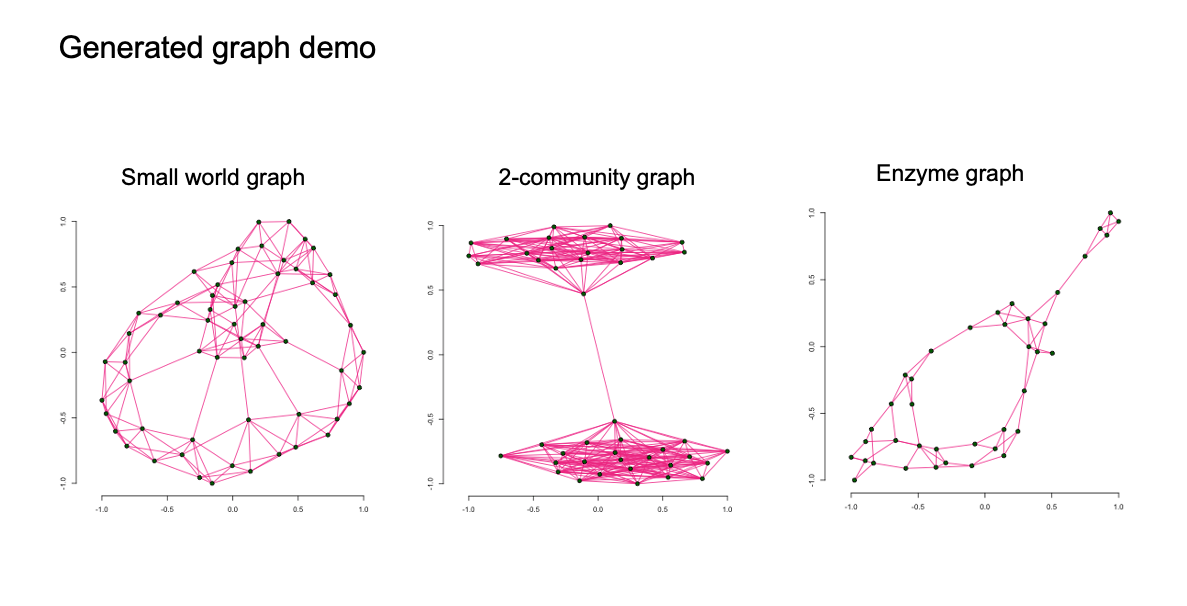
We build a graph generator that can apply to small graphs and achieve state of the art accuracy on graph generation.